Biological systems
Biological systems is an interdisciplinary research group working at the boundaries of computer science and biology. We develop novel computational and mathematical approaches to understand and design complex biological systems in the areas of synthetic, systems biology and bioinformatics.
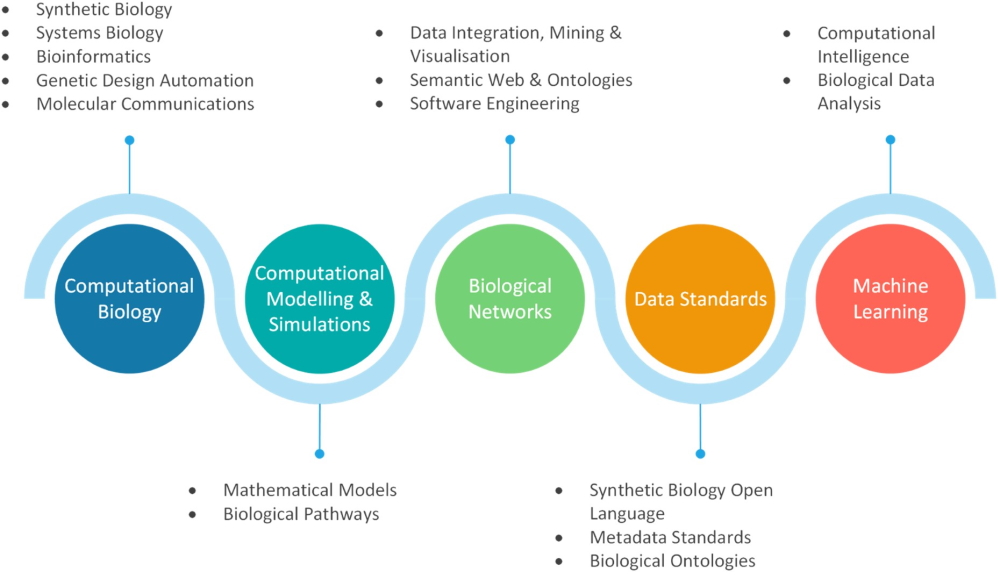
Computational biology and modelling
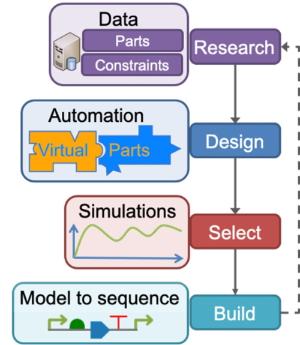 We are particularly interested in genetic design automation and the application of computational approaches in synthetic biology, involving engineering design principles such as abstraction, standardisation and modularity to facilitate the development of novel applications. Our research in computational methodologies facilitates the design of large-scale and predictable applications.
We are particularly interested in genetic design automation and the application of computational approaches in synthetic biology, involving engineering design principles such as abstraction, standardisation and modularity to facilitate the development of novel applications. Our research in computational methodologies facilitates the design of large-scale and predictable applications.
One of our strengths is developing model-driven design methodologies and creating composable and molecular models of biological components to derive predictable physical systems. Our framework to access and use these models is Virtual Parts Repository, and it can be accessed at virtualparts.org. One of our papers demonstrating the model-driven design of biological systems was selected as the best paper at the IEEE International Conference on Computational Intelligence in Bioinformatics and Computational Biology in 2014.
Biological networks and data standards
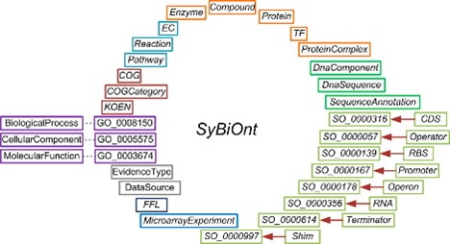 We research the development of knowledge representation techniques to utilise big data. Our research in this area includes the bridging Semantic Web technologies and the representation of biological data. We apply ontological data integration and mining methodologies to infer information.
We research the development of knowledge representation techniques to utilise big data. Our research in this area includes the bridging Semantic Web technologies and the representation of biological data. We apply ontological data integration and mining methodologies to infer information.
We extend modelling frameworks to enhance the representation and mining of biological information in computational models. Some of our work involves extending the Systems Biology Markup Language (SBML) and rule-based mathematical models to enable further developments such as computational composition, verification and understanding of mathematical models. We work with computational modelling communities in this area, e.g., the Computational Modelling in Biology (COMBINE) and contribute to the metadata standards (e.g., OMEX).
Related to this, we develop domain-specific languages to make programming for biology and facilitate information exchange easily.
SBOL
 We contribute to the development of the Synthetic Biology Open Language (SBOL) data standard, which has emerged to electronically capture information about genetic circuits and is being adopted worldwide. In addition to the SBOL specification, we develop and maintain computational frameworks and utilities to increase SBOL’s adoption.
We contribute to the development of the Synthetic Biology Open Language (SBOL) data standard, which has emerged to electronically capture information about genetic circuits and is being adopted worldwide. In addition to the SBOL specification, we develop and maintain computational frameworks and utilities to increase SBOL’s adoption.
SBOL-OWL
Our group maintains and extends the SBOL ontology, called SBOL-OWL. This ontology makes the SBOL specification available for machine access and provides a semantic reasoning layer for biological design information.
JSBOL
We also contribute to the SBOL libraries. Currently, we are developing the JSBOL Java Library for SBOL version 3. We apply software engineering and graph theory to develop related digital infrastructure.
SBOL-visual
 We are also involved in the development of SBOL Visual standard, which provides a set of glyphs to standardise the representation of genetic circuit designs.
We are also involved in the development of SBOL Visual standard, which provides a set of glyphs to standardise the representation of genetic circuit designs.
SBOL-visual ontology and the web service
We developed the SBOL-Visual Ontology. As the standard evolves and the properties of SBOL glyphs are enhanced, we capture the metadata via this ontology for machine access. The Visual Ontology Web Service (VOWS) that we developed makes the interaction of the tools with glyph metadata easier and provides a data integration mechanism with other biological ontologies.
Machine learning and computational intelligence
We are interested in applying machine learning to biological data and using computational intelligence to design biological systems. Our collaborations include modelling and tuning the fitness landscapes of biological systems via computational simulations.
Contact

Dr Charles Day
Lecturer
Computer Science
- Colin Reeves, CR110
- +44 (0)1782 733411
- c.r.day@keele.ac.uk

Dr Goksel Misirli
Senior Lecturer
Computer Science
- Colin Reeves, CR40
- +44 (0)1782 734028
- g.misirli@keele.ac.uk

